Default Homeostatic Mechanism
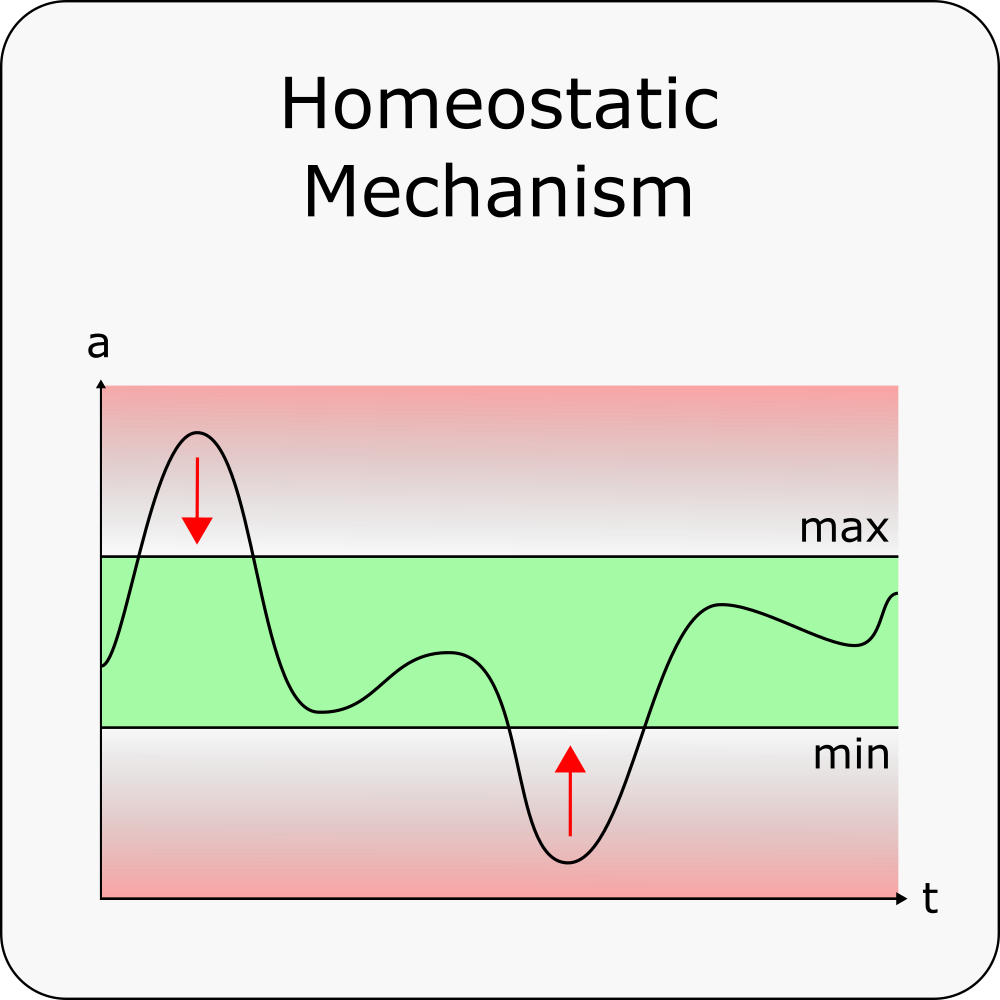
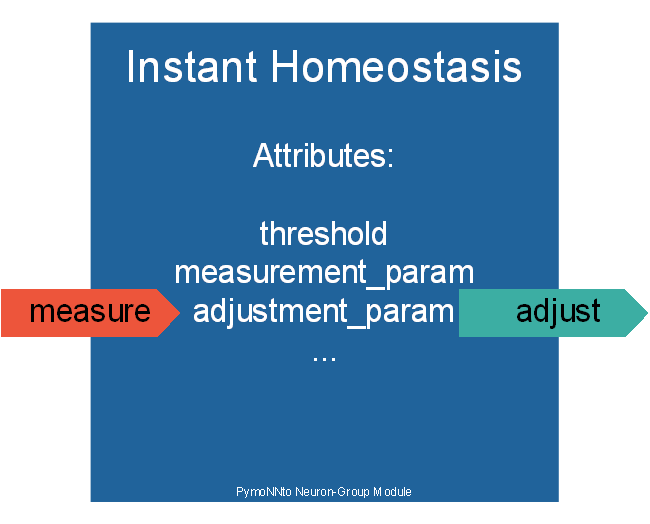
This Homeostatic Mechanism is a more general version of the homeostatic mechanism used in the Complex Tutorial. Here we also have some variable(measurement_param), which is used to determine weather a second variable(adjustment_param) has to get bigger or smaller. We also have single threshold or a min and max value to determine the target of our measurement_param. The min and max value can also be set with the two values threshold and gap_percent so that min=th(100%-gp) and max=th(100%+gp).
measurement_param
adjustment_param
min_th
max_th
threshold
gap_percent
Instant_Homeostasis(measurement_param='n.voltage', adjustment_param='n.adjust', threshold=0.5)
By default, the adjustment is distance sensitive, so the adjustment_param changes faster the further away the measurement_param variable is from the target activity. If we want a binary increase or decrease, we can set the distance_sensitive attribute to False.
The increase and decrease speed, as well as the overall speed of the adjustment can be set with the inc, dec, and adj_strength attributes. All of them are 1 by default. The next code block shows a configuration, where the increase (0.016=0.06) is faster than the decrease (0.010.5=0.005). In some cases we want the adjustment_param to get bigger when the measurement_param is above a certain value and vice versa. In this case we only have to set adj_strength to a negative value.
distance_sensitive
inc
dec
adj_strength
The target variable can also be clipped so it stays in a specific range to avoid a runaway behaviour. The measurement variable has also some clipping attributes, but this will only clip the readout, so the variable itself will not be changed.
target_clip_min
target_clip_max
measurement_min
measurement_max
Instant_Homeostasis(..., distance_sensitive=False, inc=6, dec=0.5, adj_strength=0.01, ...)
A useful function is the addition of functions to the measurement_param. Because the string is evaluated at runtime, we can also use all kinds of functions that output a value or a vector with the size of the NeuronGroup. In this Example we use the average voltage of the NeuronGroup as a measurement param but more complex functions can be used as well.
Instant_Homeostasis(..., measurement_param='np.mean(n.voltage)')
Time Integrated Default Homeostatic Mechanism
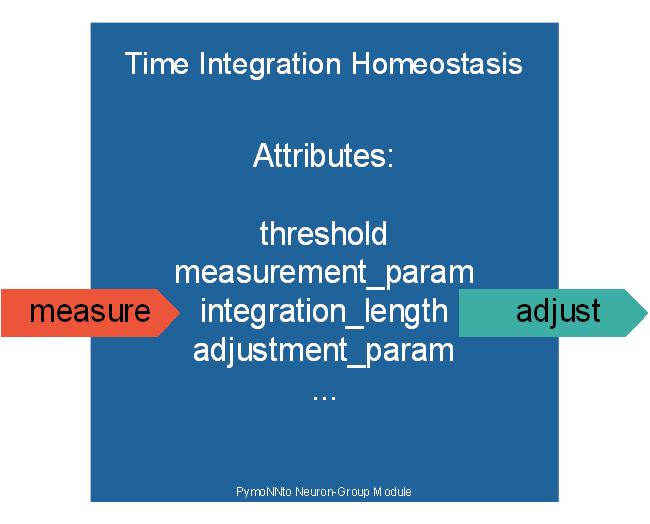
A derived version of the Default Homeostatic Mechanism is the Time Integrated Default Homeostatic Mechanism. It has the same attributes as the parent class as well as two additional parameters.
Here the measurement_param is not used directly. The value(s) are first added to an approximated average counter, which is then used for the further processing. The update of the Exponential moving average counter is the following:
a(t+1)=(a(t)*i+m)/(i+1)
where "a" is the average counter variable, "i" is the integration_length and "m" is the measurement_param.
By this, we can use the approximated average values to update the adjustment values, which are more stable for the cost of some delay. The init_avg value is the initial value of the average counter, which is (min_th+max_th)/2 by default.
integration_length
init_avg
Time_Integration_Homeostasis(..., integration_length=10)